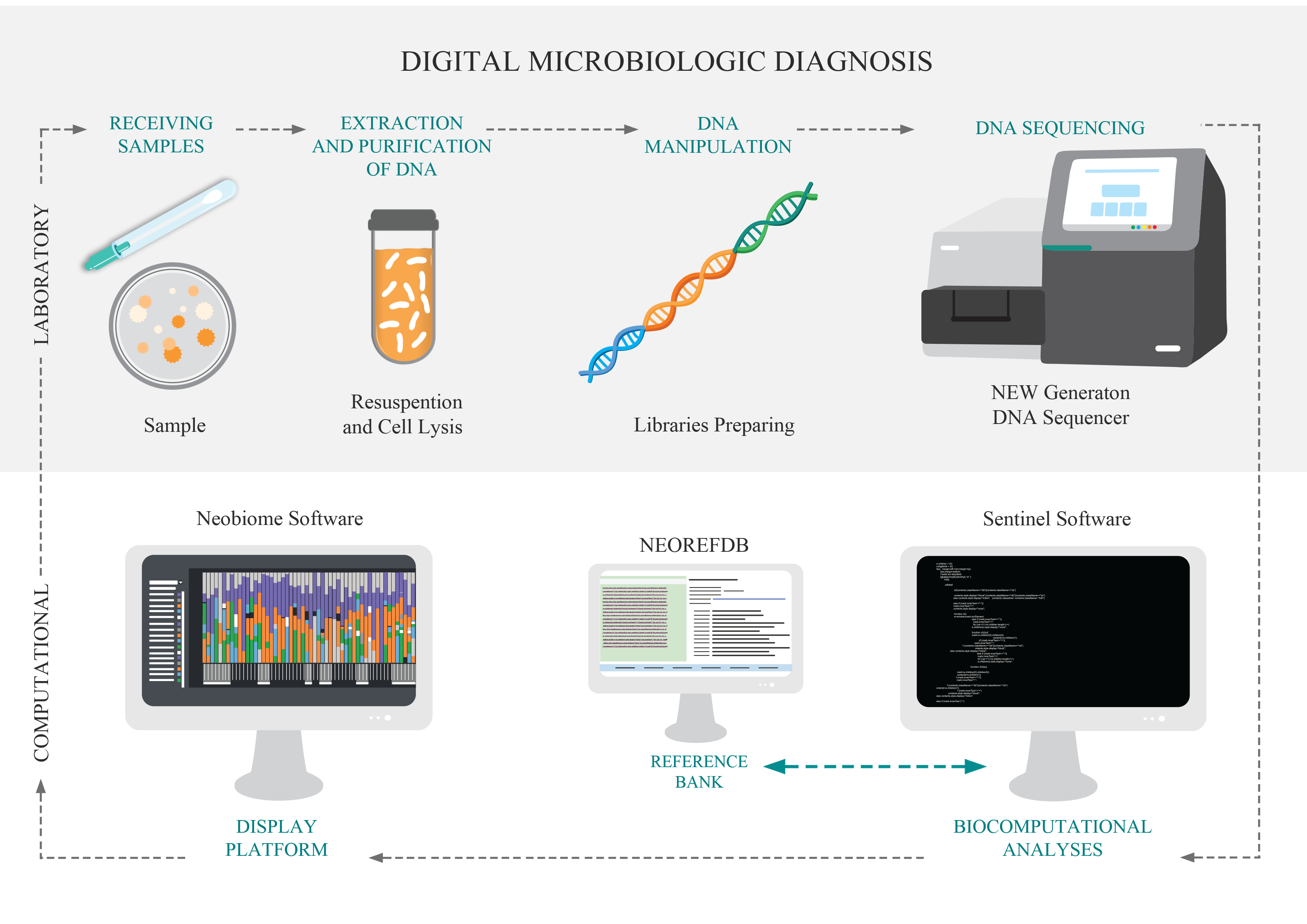
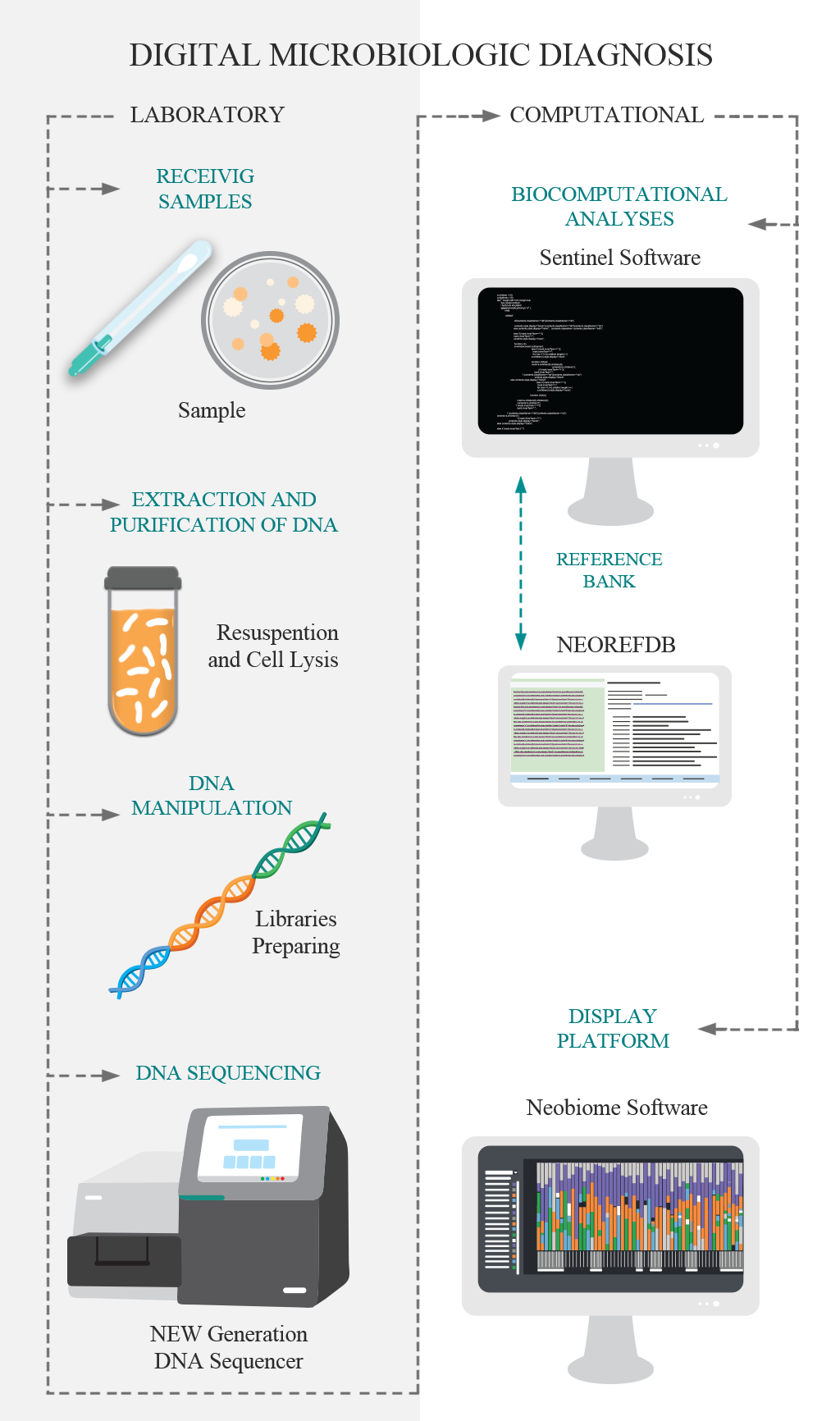
Traditional methods of microbiology have the ability to identify one microorganism at a time (at different taxonomic levels), using culture steps. The DMD, Digital Microbiological Diagnostic, was developed for breaking this paradigm, identifying the microorganisms directly from the sample using its DNA. So, the culture stage is unnecessary the culture stage. DNA is the genetic material that defines the individual, and when analyzed in a comparative way, it allows the identification of this individual at different levels. DMD is based on the use of genetic markers for the identification of microorganisms, based on the new technologies of large-scale DNA sequencing. This technological pipeline has two main stages: i. The laboratory step comprises the purification and extraction of DNA, molecular preparation of the sample and DNA sequencing; and; II) The computational steps that consist of bioinformatics analysis and the implementation of the data in the visualization platform.
• DMD | Microbiome: Pipeline for identification and quantification of fungi, bacteria and protozoa;
• DMD | Resistome: Pipeline for identification of antibiotic resistance genes;
• DMD | Virome: Pipeline for virus identification and quantification;
• DMD | HMLST: Pipeline for clonal group analysis of bacteria, using internationally standardized gene schemes;
• DMD | SalmonellaID: Pipeline for the identification of serovarieties and clonal group analysis of Salmonella.
• DMD | ViableCellsID: Identification pipeline only from viable cells DNA.
The Neobiome platform is a software developed by Neoprospecta that acts as an analytical interface for the results of the digital microbiological diagnostic. This platform allows intuitive visualization and analysis of results through innovative tools such as microbiological profile, genetic profile and risk map.
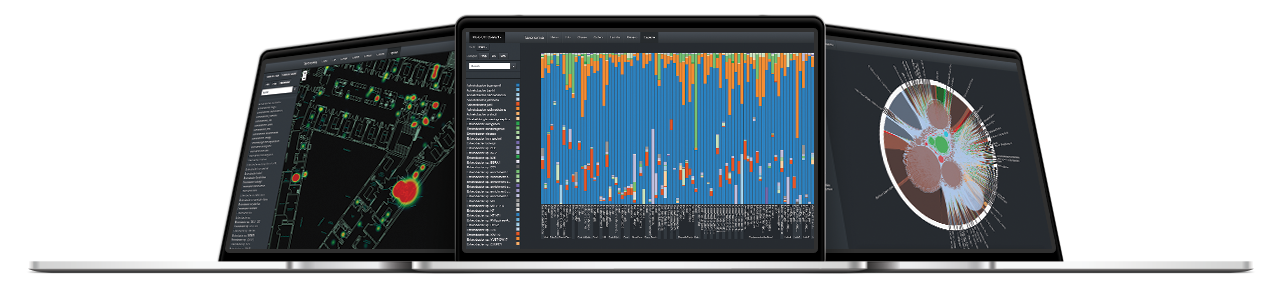
Using an algorithm that calculates risk patterns, this tool visually indicates, in a ground plan, the patterns of microbiological contaminations.
It allows visualizing the composition and quantitative relations of the microorganisms identified.
This tool allows tracing possible contamination routes by grouping together all genetically identical DNA sequences and showing in which samples this specific DNA was present.
Tell us a little about your interests
Fully automated pipeline assembly, annotation and SNP calling. This pipeline allows rapid characterization according to the NCBI Prokaryotic Genome Annotation Standards.
NEORefDB is a proprietary Neoprospecta database with great accuracy for 16S (bacteria), 18S (protozoa), ITS (Fungi), Antibiotic Resistance Genes and viruses (specific primers). This database is the base of reference used in the pipelines of the digital microbiological diagnostic.
One of the specialties of Neoprospecta is the development of kits for fast diagnostic by qPCR. With this specialty came the XGene, a line of fast diagnostic kits for identification of microorganisms and antimicrobial resistance, aimed at different segments, such as:
Hand hygiene is one of the main factors for the reduction of microbiological contaminations and outbreaks of hospital infections. With the request of clients and partners, Neoprospecta decided to develop a revolutionary approach to solve this problem. Using as assumptions the precision in the identification of the individual and the noninterference in the routine of health professionals, Neoprospecta, in partnership with SENAI / SC, developed a technology that identifies if an individual sanitized the hands, using recognition of the vein pattern of the patient’s hand palm. This module will work in any automatic dispenser, allowing its implementation with a low investment cost, at the same time, it will not have any interference in the routine of professionals. This technology can be applied in hospitals, clinics, the food industry, food services and other institutions where hand hygiene is critical to the quality of the activities.